cis_regulatory_module (CURRENT_RELEASE) |
|
|---|---|
| SO Accession: | SO:0000727 (SOWiki) |
| Definition: | A regulatory region where transcription factor binding sites are clustered to regulate various aspects of transcription activities. (CRMs can be located a few kb to hundreds of kb upstream of the core promoter, in the coding sequence, within introns, or in the untranslated regions (UTR) sequences, and even on a different chromosome). A single gene can be regulated by multiple CRMs to give precise control of its spatial and temporal expression. CRMs function as nodes in large, intertwined regulatory network. CRM DNA accessibility is subject to regulation by dbTFs and transcription co-TFs. |
| Synonyms: | cis regulatory module, CRM, TF module, transcription factor module |
| DB Xrefs: | PMID: 19660565 |
| SO: SG | |
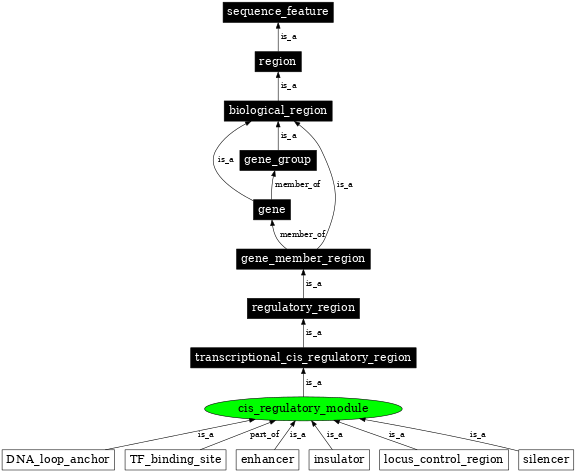
| Parent: | transcriptional_cis_regulatory_region (SO:0001055) |
| Children: | DNA_loop_anchor (SO:0002308) |
| TF_binding_site (SO:0000235) | |
| silencer (SO:0000625) | |
| locus_control_region (SO:0000037) | |
| insulator (SO:0000627) | |
| enhancer (SO:0000165) | |
In the image below graph nodes link to the appropriate terms. Clicking the image background will toggle the image between large and small formats.